This web site is currently a work in progress, but we hope that over time it will be developed into a comprehensive resource for the study of transcriptional repression in plants. If you find any errors or omissions, please contact Barry Causier. PlaCID is a description of the protein interaction partners of plant co-repressor proteins, provided by the Prof. Brendan Davies [e-mail] [www] lab and maintained by Dr. Barry Causier [e-mail].
Transcriptional repression is an important mechanism by which genes can be regulated. This database stems from our investigations into Arabidopsis TOPLESS (TPL) and the four TPL-related proteins (TPR1-4), which belong to the Gro/Tup1 class of co-repressors first identified in Drosophila and yeast, respectively. TPL/TPR proteins interact with a broad range of transcription factor families, many of which are transcriptional repressors, with roles in diverse developmental processes including hormone signalling and stress responses.
We recently published the interactome for the TPL/TPR proteins (Causier et al. (2012) Plant Physiol. 158: 423). Here we present that interactome, together with previously published TPL interactions from various species. We also list reported interactions for other plant co-repressors including LUG/LUH, SEU/SLK, the LUG-SEU complex and SAP18 family proteins.
The TPL/TPR interactome (Causier et al., 2012) was established through two large-scale yeast two-hybrid approaches - whole plant library screens and screens of an arrayed library of Arabidopsis transcription factors:
Whole plant library screens
| AGI | Identity | TPL | TPR1 | TPR2 | TPR3 | TPR4 | RDs | ||
| AT3G02550 | LBD41 | LDLTLRL | |||||||
| AT5G67420 | LBD37 | LDLSL | |||||||
| AT1G50640 | ERF3 | AP2/ERF | FDLNFPP | * | |||||
| AT3G15210 | ERF4 | AP2/ERF | LELSL/LDLNLPP | * | |||||
| AT3G20310 | ERF7 | AP2/ERF | FDLNFPP | * | |||||
| AT1G53170 | ERF8 | AP2/ERF | LELSL/LDLDLNLAP | * | |||||
| AT1G28370 | ERF11 | AP2/ERF | LDLDLNFPP | * | |||||
| AT1G18330 | EPR1 | MYB | partial | ||||||
| AT2G16720 | MYB7 | MYB | LNLEL | ||||||
| AT4G34990 | MYB32 | MYB | LDLNLEL | ||||||
| AT5G67300 | MYB44 | MYB | LSLSL | ||||||
| AT3G50060 | MYB77 | MYB | LSLSL | ||||||
| AT3G52250 | MYB/HD | MYB | partial | ||||||
| AT5G17300 | MYB (EPR-like) | MYB | LKLELFP | * | |||||
| AT1G46480 | WOX4 | HD | TLELFPL | ||||||
| AT1G27730 | ZAT10 | ZnF | FDLNIPP | ||||||
| AT5G04340 | ZAT6 | ZnF | LCLML/DLNIPP | ||||||
| AT1G08290 | WIP3 | ZnF | partial | ||||||
| AT2G42410 | ZFP11 | ZnF | LDLELRL | ||||||
| AT1G30810 | Zn finger TF | ZnF | KLFGV/FDLNIDL | ||||||
| AT2G28200 | ZAT5 | ZnF | LPLDLNLP | ||||||
| AT2G27110 | FRS3 | ZnF | DLNLP | ||||||
| AT3G50870 | MONOPOLE/GATA18 | ZnF | partial | ||||||
| AT1G17380 | JAZ5 | DLNEPT/LDLRL | |||||||
| AT1G72450 | JAZ6 | DLNEPT/LELKL | |||||||
| AT3G23030 | IAA2 | AUX/IAA | LCLGL | * | |||||
| AT3G23050 | IAA7/AXR2 | AUX/IAA | LCLGL/LMLNL | * | |||||
| AT2G22670 | IAA8 | AUX/IAA | LRLGL | ||||||
| AT3G04730 | IAA16 | AUX/IAA | LRLGL/LKLNL | ||||||
| AT1G51950 | IAA18 | AUX/IAA | LELKL | ||||||
| AT4G29080 | IAA27 | AUX/IAA | LRLGL | ||||||
| AT1G04250 | AXR3 | AUX/IAA | LCLGL/LKLNL | ||||||
| AT5G62000 | ARF2 | ARF | LSLKL/RLFGI | ||||||
| AT4G23980 | ARF9 | ARF | RLFGI | ||||||
| AT3G61830 | ARF18 | ARF | RLFGF | ||||||
| AT1G25560 | TEM1 | AP2/ERF | RLFGV | ||||||
| AT2G28550 | TOE1 | AP2/ERF | LDLNL | * | |||||
| AT5G60120 | TOE2 | AP2/ERF | LDLNL/LDLNL | * | |||||
| AT2G36080 | B3 TF | RLFGV | |||||||
| AT5G55040 | BROMO TF | LELIL | |||||||
| AT1G58100 | TCP8 | TCP | partial | * | |||||
| AT3G63180 | TKL | partial | |||||||
| AT1G78700 | BEH4 | LELTL | |||||||
| AT4G36990 | HSF4 | KLFGV | |||||||
| AT2G47070 | SPL1 | LTLNL | * | ||||||
| AT1G80490 | TPR1 | partial | |||||||
| AT3G15880 | TPR4 | none | |||||||
| AT5G44800 | PKR1 | LPLFL | |||||||
| AT3G16840 | HELICASE | partial | |||||||
| AT3G29575 | AFP3 | LDLGLSL | |||||||
| AT3G02140 | AFP4 | LTLGLSL | |||||||
| AT2G19520 | FVE | partial | |||||||
| AT5G11530 | EMF1 | partial | |||||||
| AT3G24440 | VRN5 | partial | |||||||
| AT3G44630 | TIR-NBS-LRR | partial | |||||||
| AT4G34370 | ATARI1 | LDLNLP | |||||||
| AT5G55170 | SUMO3 | partial | |||||||
| AT4G32030 | unknown | LLLRL/DLNMAP | |||||||
| AT1G12120 | unknown | DLNSTP | |||||||
| AT1G80610 | unknown | DLNMP | |||||||
| AT5G57340 | hypothetical | LELTL | |||||||
| AT4G32295 | unknown | LSLRL | |||||||
| AT5G42530 | unknown | LFLLLLL | |||||||
| AT5G14970 | unknown | partial | |||||||
| AT3G17900 | unknown | LELLPL |
Arrayed TF library screens
| AGI | Identity | TF | TPL | TPR1 | TPR2 | TPR3 | TPR4 | RDs | |
| AT2G28550 | TOE1 | AP2/ERF | LDLNL | * | |||||
| AT1G04100 | IAA10 | AUX/IAA | LDLALGL | ||||||
| AT3G23030 | IAA2 | AUX/IAA | LCLGL | * | |||||
| AT1G77850 | ARF17 | ARF | partial | ||||||
| AT5G60120 | TOE2 | AP2/ERF | LDLNL/LDLNL | * | |||||
| AT4G14560 | IAA1 | AUX/IAA | LRLGL | ||||||
| AT4G28640 | IAA11 | AUX/IAA | LELGLTLSL | ||||||
| AT2G37630 | AS1 | MYB | LELQL | ||||||
| AT5G24800 | BZIP9 | bZIP | partial | ||||||
| AT2G16770 | BZIP23 | bZIP | none | ||||||
| AT1G13260 | RAV1 | AP2/ERF | RLFGV | ||||||
| AT2G47070 | SPL1 | SBP | LTLNL | * | |||||
| AT3G11580 | AP2/B3 | AP2/ERF | RLFGV | ||||||
| AT3G15540 | IAA19 | AUX/IAA | LRLGL | ||||||
| AT4G11660 | HSFB2B | HSF | RLFGV | ||||||
| AT1G51120 | AP2/B3 | AP2/ERF | MLFGV | ||||||
| AT4G36920 | AP2 | AP2/ERF | DLNDAP/LDLSL | ||||||
| AT4G00540 | MYB3R2 | MYB | LDLTLGL | ||||||
| AT3G15030 | TCP4 | TCP | LRLSL | ||||||
| AT2G45660 | SOC1 | MADS | partial | ||||||
| AT4G37260 | MYB73 | MYB | LSLSL | ||||||
| AT2G33310 | IAA13 | AUX/IAA | LELGLGLSL | ||||||
| AT1G68670 | MYB family | MYB | LPLCLEL | ||||||
| AT2G33550 | GT-2 RELATED | MYB | LSLGL/LNLKL | ||||||
| AT5G13790 | AGL15 | MADS | LQLGL | ||||||
| AT2G37740 | ZFP10 | ZnF | LDLELRL | ||||||
| AT5G17300 | RVE1 | MYB | LKLELFP | * | |||||
| AT1G66140 | ZFP4 | ZnF | LNLSL/LTLPL/LTLKL | ||||||
| AT1G04240 | IAA3/SHY2 | AUX/IAA | LRLGL | ||||||
| AT5G43700 | IAA4 | AUX/IAA | LRLGL | ||||||
| AT5G49300 | GATA16 | ZnF | none | ||||||
| AT1G50680 | AP2/B3 | AP2/ERF | MLFGV | ||||||
| AT3G20310 | ERF7 | AP2/ERF | FDLNFPP | * | |||||
| AT1G50640 | ERF3 | AP2/ERF | FDLNFPP | * | |||||
| AT4G28530 | NAC074 | NAC | partial | ||||||
| AT1G53230 | TCP3 | TCP | LRLSL | ||||||
| AT4G35610 | Zn finger | ZnF | DLNVEP/FDLNKSP | ||||||
| AT4G17460 | HAT1 | HD | LGLSLSL/LQLNL | ||||||
| AT1G28360 | ERF12 | AP2/ERF | LSLDLNHLP/DLNEPP | ||||||
| AT2G38300 | TF (G2-like) | MYB | LDLSLKL/LSLSL/LDLTL | ||||||
| AT3G15210 | ERF4 | AP2/ERF | LELSL/LDLNLPP | * | |||||
| AT3G10490 | NAC51/52 | NAC | partial | ||||||
| AT3G10480 | NAC50 | NAC | partial | ||||||
| AT4G18390 | TCP2 | TCP | partial | ||||||
| AT5G59340 | WOX2 | HD | TLPLFP | ||||||
| AT2G44410 | Zn binding | ZnF | LDLNQEP | ||||||
| AT2G31720 | TF (B3) | LDLELRL | |||||||
| AT4G37790 | HAT22 | HD | LVLGLGL/LTLSL | ||||||
| AT3G16500 | IAA26/PAP1 | AUX/IAA | LELRL | ||||||
| AT5G25890 | IAA28 | AUX/IAA | LELRL | ||||||
| AT1G03800 | ERF10 | AP2/ERF | DLNASP | ||||||
| AT1G73100 | SDG19 | partial | |||||||
| AT2G41940 | ZFP8 | ZnF | LDLHL | ||||||
| AT1G28370 | ERF11 | AP2/ERF | LDLDLNFPP | * | |||||
| AT1G53170 | ERF8 | AP2/ERF | LELSL/LDLDLNLAP | * | |||||
| AT3G61970 | NGA2 | AP2/ERF | RLFGV/LQLRL | ||||||
| AT5G16560 | KAN | MYB | partial | ||||||
| AT5G06250 | AP2/B3 | AP2/ERF | RLFGV | ||||||
| AT5G46590 | NAC096 | NAC | partial | ||||||
| AT1G18860 | WRKY61 | WRKY | partial | ||||||
| AT3G12730 | MYB family | MYB | partial | ||||||
| AT4G03250 | HD-ZIP | none | |||||||
| AT5G63470 | NF-YC4 | partial | |||||||
| AT1G08320 | bZIP21 | bZIP | partial | ||||||
| AT5G51990 | CBF4 | AP2/ERF | partial | ||||||
| AT4G32890 | GATA9 | ZnF | partial | ||||||
| AT4G30935 | WRKY32 | WRKY | partial | ||||||
| AT5G06960 | OBF5 | bZIP | partial | ||||||
| AT1G62300 | WRKY6 | WRKY | partial | ||||||
| AT3G44460 | DPBF2/bZIP67 | bZIP | LSLTL/LELEL | ||||||
| AT1G25550 | MYB family | MYB | LPLCLEL | ||||||
| AT3G15500 | NAC3/NAC55 | NAC | partial | ||||||
| AT3G45150 | TCP16 | TCP | partial | ||||||
| AT1G35560 | TCP family | TCP | partial | ||||||
| AT4G26930 | MYB97 | MYB | partial | ||||||
| AT3G12250 | TGA6/bZIP45 | bZIP | partial | ||||||
| AT1G71030 | MYBL2 | MYB | LILKL/TLLLFR | ||||||
| AT1G66230 | MYB20 | MYB | LDLLL | ||||||
| AT3G23250 | MYB15 | MYB | partial | ||||||
| AT5G24110 | WRKY30 | WRKY | none | ||||||
| AT3G23050 | IAA7/AXR2 | AUX/IAA | LCLGL/LMLNL | * | |||||
| AT3G16350 | MYB family | MYB | KLFGV/LELSL/LSLRL | ||||||
| AT1G58100 | TCP8 | TCP | partial | * | |||||
| AT5G62020 | HSFB2A | HSF | RLFGV | ||||||
| AT3G47620 | TCP14 | TCP | partial | ||||||
| AT3G21890 | Zn finger | ZnF | none | ||||||
| AT4G18770 | MYB98 | MYB | none | ||||||
| AT5G17430 | BABYBOOM | AP2/ERF | partial |
In each of the tables above, the first column provides the AGI number for each factor. The second column indicates the identity of the factor, if known. Factors highlighted in grey in this column represent those that are known to act as transcriptional repressors. The third column indicates the transcription factor family to which each factor belongs. Interactions between these factors and the TPL/TPR proteins are represented by dark grey boxes in the appropriate columns. The RD column shows the sequence of any known functional repression domain found in the TPL/TPR-interacting factors. An asterix to the right-hand side of each table indicate those factors isolated in both the whole plant library and arrayed transcription factor screens.
Other published co-repressor interactions
TPL/TPR family
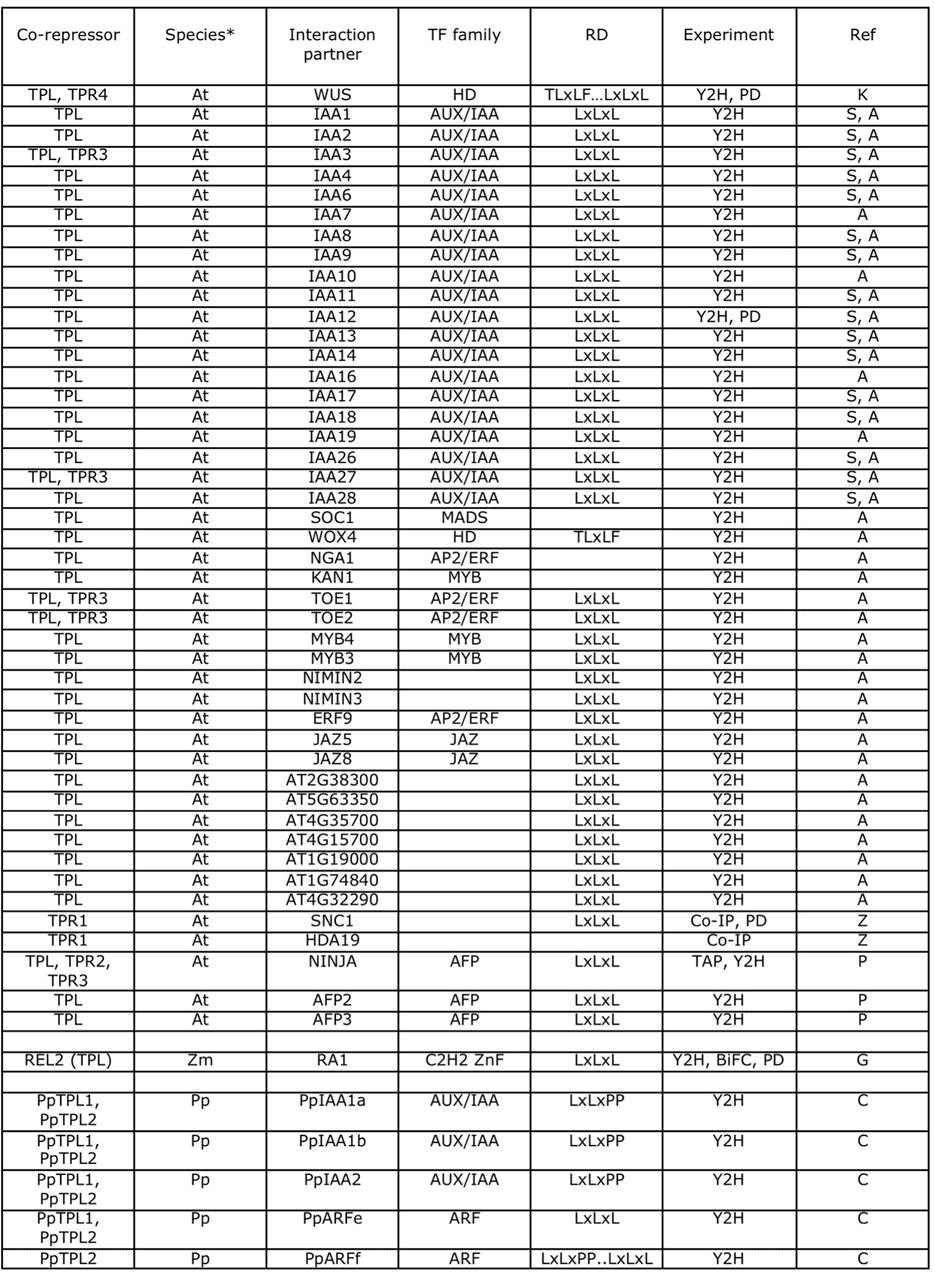
*At – Arabidopsis thaliana; Am – Antirrhinum majus; Pp – Physcomitrella patens; Zm – Zea mays
Experiments: Y2H – yeast two-hybrid; PD – pull-down; Co-IP – co-immunoprecipitation; TAP – tandem affinity purification; BiFC – biomolecular fluorescence complementation
Refs:
- A - Arabidopsis Interactome Mapping Consortium (2011) Science 333: 601.
- C – Causier et al. (2012) Plant Signalling and Behavior 7: in press
- G – Gallavotti et al. (2010) Development 137: 2849.
- K – Kieffer et al. (2006) Plant Cell 18: 560.
- P – Pauwels et al. (2010) Nature 464: 788.
- S – Szemenyei et al. (2008) Science 319: 1384.
- Z – Zhu et al. (2010) PNAS 107: 13960.
LUG/LUH family

*At – Arabidopsis; Am - Antirrhinum
Experiments: Y2H – yeast two-hybrid; PD – pull-down; BRET – bioluminescence resonance energy transfer Refs:
- G – Gregis et al. (2006) Plant Cell 18: 1373.
- N – Navarro et al. (2004) Development 131: 3649.
- St – Stahle et al. (2009) Plant Cell 21: 3105.
- S1 – Sridhar et al. (2004) PNAS 101: 11494.
SEU/SLK family
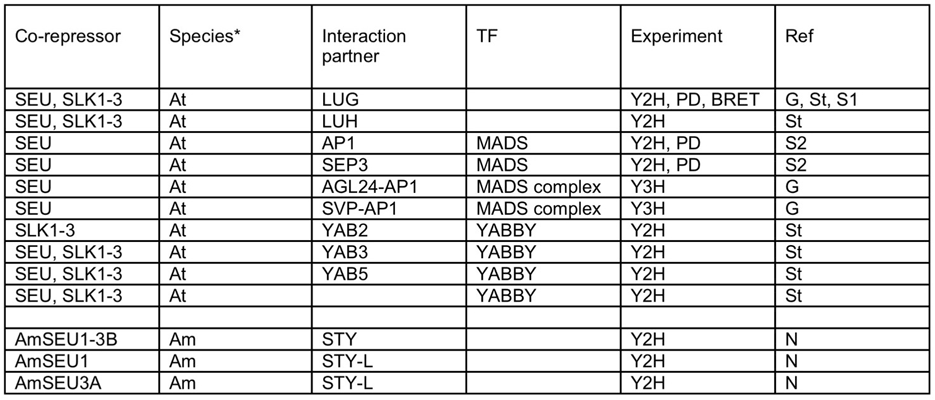
*At – Arabidopsis; Am – Antirrhinum
Experiment: Y2H – yeast two-hybrid; Y3H – yeast three-hybrid; PD – pull-down; BRET – bioluminescence resonance energy transfer
Refs:
- G – Gregis et al. (2006) Plant Cell 18: 1373.
- N – Navarro et al. (2004) Development 131: 3649.
- St – Stahle et al. (2009) Plant Cell 21: 3105.
- S1 – Sridhar et al. (2004) PNAS 101: 11494.
- S2 – Sridhar et al. (2006) Development 133: 3159.
LUG/SEU complexes

*At – Arabidopsis; Am – Antirrhinum
Experiment: Y4H – yeast four-hybrid; Y3H – yeast three-hybrid; PD – pull-down
Refs:
- G – Gregis et al. (2006) Plant Cell 18: 1373.
- N – Navarro et al. (2004) Development 131: 3649.
- S2 – Sridhar et al. (2006) Development 133: 3159.
Arabidopsis SAP18
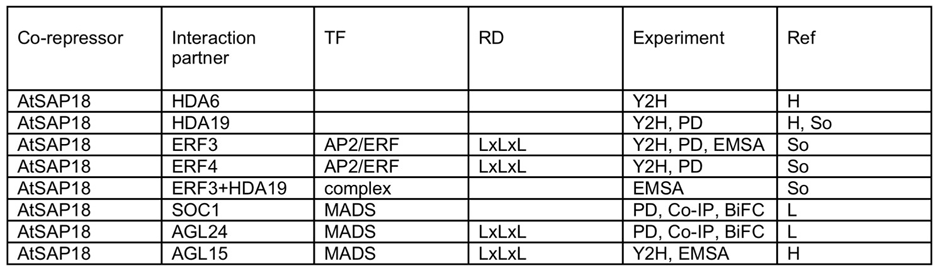
Experiment: Y2H – yeast two-hybrid; PD – pull-down; EMSA – electrophoretic mobility shift assay; Co-IP – co-immunoprecipitation; BiFC – biomolecular fluorescence complementation
Refs:
- H – Hill et al. (2008) Plant J 53: 172.
- So – Song & Galbraith (2006) Plant Mol Biol 60: 241.
- L – Liu et al. (2009) Dev Cell 16: 711.
